About the Bangor Imaging Unit
The Bangor Imaging Unit (BIU) is a cutting-edge hub for discovery and innovation in neuroscience and human physiology. Equipped with research and teaching dedicated brain imaging and brain stimulation laboratories, it is geared to unlock the mysteries of human biology. Open to researchers and students alike, it facilitates discoveries in science and medicine, and training of the next generation of knowledge pioneers.
Established in 2006 with support from the Welsh Government and the Wolfson Foundation, the BIU is supported by several full-time staff and faculty. Our academics have generated funding from a wide range of sources and regularly publish in international peer-reviewed journals.
For the latest round-up of information and news, please read our most recent Annual Report below:
Discovery and Innovation
Research Themes
Research into human physiology is unlocking the secrets of life, transforming knowledge into innovations that heal, uplift, and inspire humanity.
We seek to understand the structural, chemical and functional composition of the human body and all its systems, with the aim of understanding what constitutes healthy versus diseased or disordered states, and how we can work to prevent and repair harm.
Our researchers are actively studying the interplay between brain activity, connectivity, and the neurochemical and physiological processes underlying neural function and plasticity.
We do this by working with individuals with a variety of conditions, including participants who have brain injury, those who have undergone limb amputation or injury, and those with social difficulties and/or challenges with speech/language. In addition, we investigate the basic physiologic effects of physical and environmental stressors like exercise, fatigue and low oxygen levels.
By exploring a wide spectrum of conditions and environmental influences, we will fulfil our goal of having a meaningful impact, enriching our community with the knowledge necessary to lead a long and healthy life.
Investigators
Our researchers are world leaders in developing novel imaging methods to enhance neuroscience discovery. This includes developing sequential proton magnetic resonance spectroscopy to investigate neurotransmitter dynamics, real-time vocal tract MRI to track speech articulation in those who stammer, and bespoke methods to improve signal-to-noise in otherwise hard-to-reach areas of the brain involved in memory, language and social cognition.
Investigators
Our brains are incredibly efficient at extracting complex information from our environment and using it to guide our actions. This makes us agile and resilient, and it underpins our ability to problem solve and be creative.
Our researchers study how we process sensory inputs, how we control our motor output, and how we retain knowledge over time. This includes how we process bodily sensations and perception of self.
We work with diverse populations, including children, adults, and older individuals, those with neurological or developmental conditions, and those with physical disabilities.
This research addresses real-world challenges such as improving rehabilitation strategies for injuries or disorders, and informing technologies like AI and human-computer interfaces. Understanding these processes fosters innovation in healthcare and everyday decision-making.
Investigators
Humans are uniquely social and interacting with one another is essential for our well-being.
We are interested in how our brain makes sense of other people, their words and their actions, and the processes underlying production of speech and language, facial expressions and bodily gestures. We also study the way in which humans interact with robots and machines.
Our researchers study social cognition and language across the lifespan, from infants and adolescents to the ageing population, as well as in the context of neurodiversity, and in brain injury and disease.
We envisage using our research findings to help people manage their language disabilities and social difficulties.
Investigators
The BIU Team
The Bangor Imaging Unit (BIU) is home to a vibrant interdisciplinary community of neuroscientists, physiologists, physicists and clinicians who engage in both fundamental and applied research. Individual Principal Investigator Profiles can be found in the drop-down menu below, and under our Research Themes.
Strategic and operational management of the BIU is the responsibility of an academic director, and a committee that includes a senior physicist and professional services staff. More information on these individuals can be found below.
Facilities
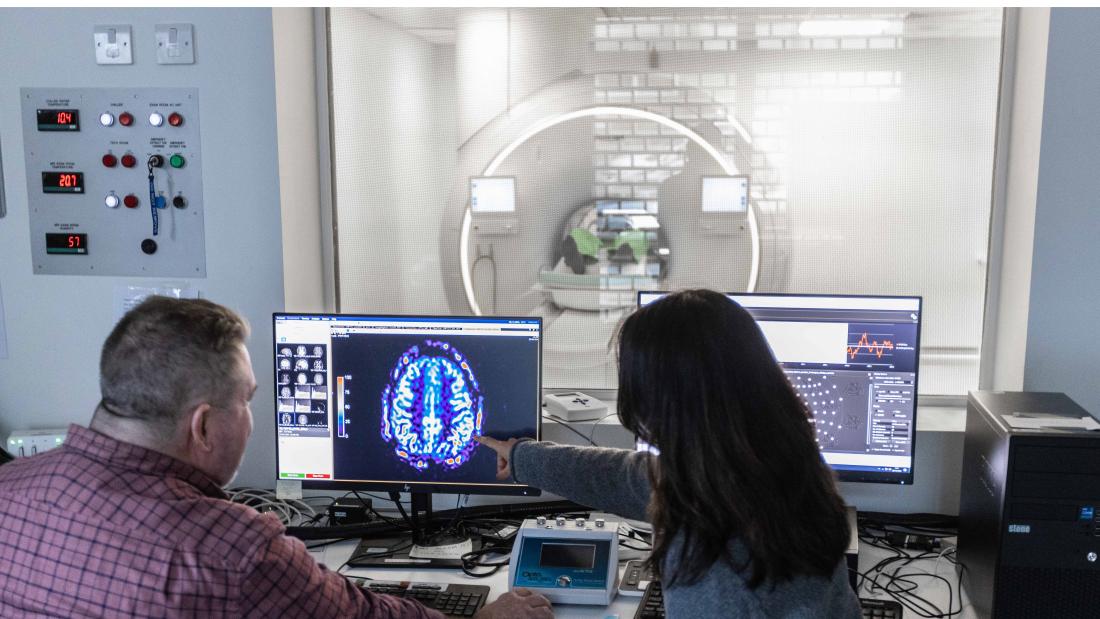
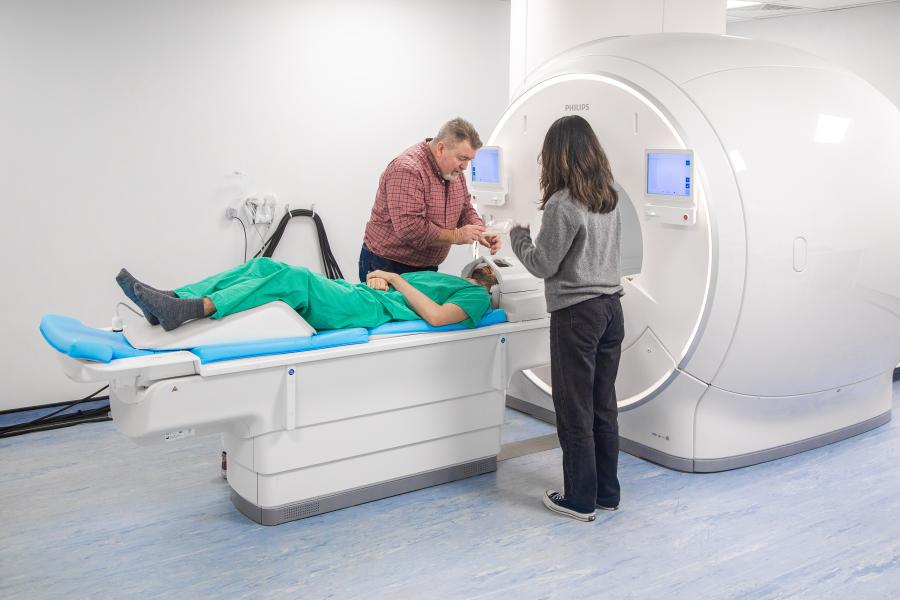
The Bangor Imaging Unit houses a Philips “Elition X” 3T MRI system, installed in July 2019. This new generation MRI full body platform enables high signal-to-noise and high-spatial resolution head-to-toe imaging. With high speed, high duty gradients, compressed SENSE, and simultaneous multiband imaging, the 3T Elition enables increased temporal resolution for functional acquisitions and overall faster imaging.
The Elition X comes with an impressive array of advanced imaging sequences, allowing high resolution mapping of structure (DTI, DSI and white matter tractography) and metabolism (arterial spin labelling for cerebral blood flow, Angiography, magnetic resonance spectroscopy (MRS), and amide proton transfer (APT)).
While we are predominantly focused on the brain and central nervous system at the BIU, we also have a full suite of musculoskeletal, cardiac, renal, and peripheral imaging techniques available, allowing other organ systems to be investigated.
Peripheral accessories include:
- specialised stimulus delivery computers
- custom built participant interface devices
- physiologic status measurements (SpO2, ETCO2, respiration, VCG)
- enhanced visual and auditory delivery systems
- custom built exercise equipment
- image analysis systems for student researchers
What can we understand from using this technique?
MRI enables research on the organisation of structure and function of complex organs such as the brain, kidneys and muscles, and how this is affected by experience, exercise, and disease.
How can researchers access this facility?
We have a well-established set of internal procedures for training researchers, for ethics and for research governance. If you are an external researcher, business or community organisation interested in learning more about, or accessing BIU facilities, please contact the unit director who will advise on the opportunities. Alternatively, you may identify a suitable collaborator from our Principal Investigator list and contact them directly with a proposal.
When using the MRI scanner to take images of the brain, it is of vital importance that participants keep their head (and body) as still as possible. The mock scanner facility is designed to familiarise participants, particularly children, with being in the scanner environment.
Our state-of-the-art mock scanner is equipped with a sound system to play scanner sounds and MoTrak, a software which allows fine-grained head motion tracking.
What can we understand from using this technique?
MRI enables research on the organisation of structure and function of complex organs such as the brain, and how this is affected by experience and disease.
How can researchers access this facility?
We have a well-established set of internal procedures for training researchers, for ethics and for research governance. If you are an external researcher, business or community organisation interested in learning more about, or accessing BIU facilities, please contact the unit director who will advise on the opportunities. Alternatively, you may identify a suitable collaborator from our Principal Investigator list and contact them directly with a proposal.
The Bangor Imaging Unit has a portable and MRI-compatible fNIRS device which allows brain blood flow and oxygenation status to be collected both in the MRI scanner, and in more naturalistic environmental settings. This allows us to explore the effects of a range of stressors including temperature, exertion, dehydration, and the impact of disease. The device also allows a range of other research avenues - neurofeedback experiments to improve task-based efficiency, monitoring of cerebral responses in pharmaco-physiology studies, and monitoring functional changes in occupational rehabilitation after acquired brain injury.
We have complementary instrumentation to manipulate and measure other aspects of basic physiology, including:
- increased exertion (MRI compatible exercise bike),
- changes in atmosphere breathed (MRI compatible gas delivery device),
- heart rate, blood pressure, and blood oxygenation (MRI compatible physiologic monitoring).
What can we understand from using this technique?
fNIRS enables research on how blood oxygenation in the brain responds to mental or physical activity and, for example, how this is affected by medication or disease. It is an alternative for exploring brain function in people or environmental contexts that are not well suited for MRI.
How can researchers access this facility?
We have a well-established set of internal procedures for training researchers, for ethics and for research governance. If you are an external researcher, business or community organisation interested in learning more about, or accessing BIU facilities, please contact the unit director who will advise on the opportunities. Alternatively, you may identify a suitable collaborator from our Principal Investigator list and contact them directly with a proposal.
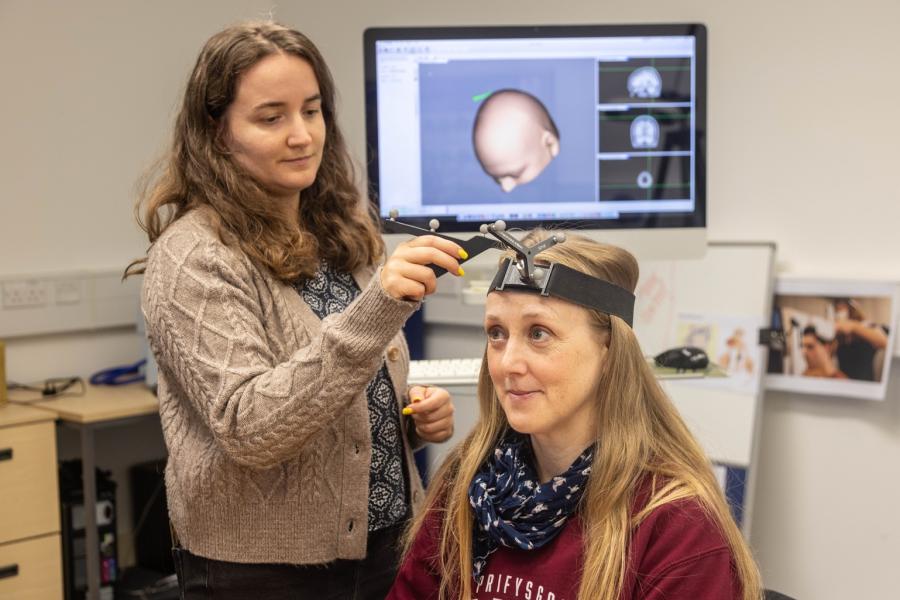
The Bangor Imaging Unit is located directly next to a brain stimulation lab, complete with multiple transcranial magnetic stimulators, transcranial direct/alternating current stimulators, and a Brainsight neuronavigation system for functional and anatomical MRI-guided brain stimulation. Our Brainsight system includes a two-channel EMG module enabling flexible visualisation and recording of motor evoked potentials (MEP). We have a wide range of TMS coils, including the Magstim D11 “cone coil” specialised for stimulation at greater depths, a cooled coil, and multiple “butterfly” coils of differing diameters and handle configurations.
Our Magstim Rapid Plus system is specialised for high-frequency repetitive TMS, useful for temporarily changing the functional ‘state’ of a brain area. This approach is used, for example, with ‘offline’ TMS experiments, where TMS is applied prior to the participant performing the task that is used to test its predicted effects.
Our Magstim Bi-Stim system is specialised for paired-pulse TMS applications and is better suited for estimating TMS-induced physiological changes, such as MEPs. These features complement our Rapid Plus system, enabling, for example, more precise and comprehensive evaluation of predicted high-frequency repetitive TMS on motor physiology.
What can we understand from using this technique?
NiBS helps us understand brain function, brain connectivity and the causal role of specific brain regions in behaviour and cognition.
How can researchers access this facility?
We have a well-established set of internal procedures for training researchers, for ethics and for research governance. If you are an external researcher, business or community organisation interested in learning more about, or accessing BIU facilities, please contact the unit director who will advise on the opportunities. Alternatively, you may identify a suitable collaborator from our Principal Investigator list and contact them directly with a proposal.
Bangor Imaging Unit, Brigantia Building, Penrallt Road, Bangor University, LL57 2AS
Contact Us
Director - Dr Richard J. Binney, School of Psychology and Sport Science
Senior Technician - Mr Andrew Fischer